Projects
admr package
Pharmacometric modelling is traditionally performed using individual level data. Recently a new method was developed to fit pharmacometric models to summary level – or aggregate – data. This methodology allows for jointly modelling different data sources, once transformed into aggregate data. As such, the method can be applied to a combination of individual data, pharmacometric models, and aggregate data. In this study we aimed to (1) implement this methodological framework into an accessible R package (admr) and (2) develop a novel algorithm with enhanced computational efficiency. The developed R-package allows calculating aggregate data from different data sources, jointly fitting one or multiple data sources and assessing model performance. The implementation of the newly developed algorithm improves computational efficiency by iteratively reweighting internal Monte Carlo predictions. Three simulation scenarios using different data generating models demonstrated an improvement of 3 to 100-fold speed-up when using the novel Iterative Reweighting Monte Carlo (IR-MC) algorithm, while maintaining the convergence properties of the original MC algorithm. These analyses demonstrated that estimation with the IR-MC algorithm is increasingly more efficient as model complexity rises as compared to the standard MC algorithm, indicating the utility for more complex pharmacometric models. In conclusion, the aggregate data modelling implementation in the admr R package allows for a fast and user-friendly application of the aggregate data modelling framework.
A preprint of the article can be found here. The package can be found on Github
Bayesian Statistics
In this project for the course Bayesian Statistics by Prof. dr. Herbert Hoijtink, the main goal was to develop a Gibbs sampler for a Bayesian linear regression model. By using two simulated data set the Gibbs sampler was tested and the results were compared. The Gibbs sampler was then applied to a real data set to identify the key factors that play a role in the University admittance process, using a open source data set. We used non-informative conjugate priors, as specified in the info note underneath. Next to this Bayesian Model Averaging was used to identify the most important variables in the model. The project was rewarded a 10.0/10.0 grade. The project can be found here.
Theorem
\(Y_i = b_0 + b_1x_{1i} + b_2x_{2i} + b_3x_{3i} + e_i\) with \(e_i\) i.i.d. \(\sim N(0,1/\tau)\) and priors
\(b_0 ... b_3 \sim N(0,1/\tau_b)\)
\(\tau \sim gamma(\alpha, \beta)\)
conditional distributions:
\(f(b_0|b_1,b_2,b_3,\tau,Y_1,...,Y_n) \sim N(\frac{\tau}{n\tau + \tau_b}\sum_{i=1}^n(Y_i -(b_1x_{1i} + b_2x_{2i} + b_3x_{3i}), \frac{1}{n\tau + \tau_b})\)
\(f(b_1|b_0,b_2,b_3,\tau,Y_1,...,Y_n) \sim N(\frac{\tau\sum_{i=1}^n(Y_i-(b_0 + b_2x_{2i} + b_3x_{3i})x_{1i})}{\tau\sum_{i=1}^n(x_i^2+\tau)}, \frac{1}{\tau\sum_{i=1}^nx_{1i}^2 + \tau_b})\)
...
\(f(\tau|b_0,b_1,b_2,b_3,Y_1,...,Y_n) \sim gamma(\alpha+n/2,\beta+\frac{1}{2}\sum_{i=1}^n(Y_i -(b_0 + b_1x_{1i} + b_2x_{2i} + b_3x_{3i})^2))\)
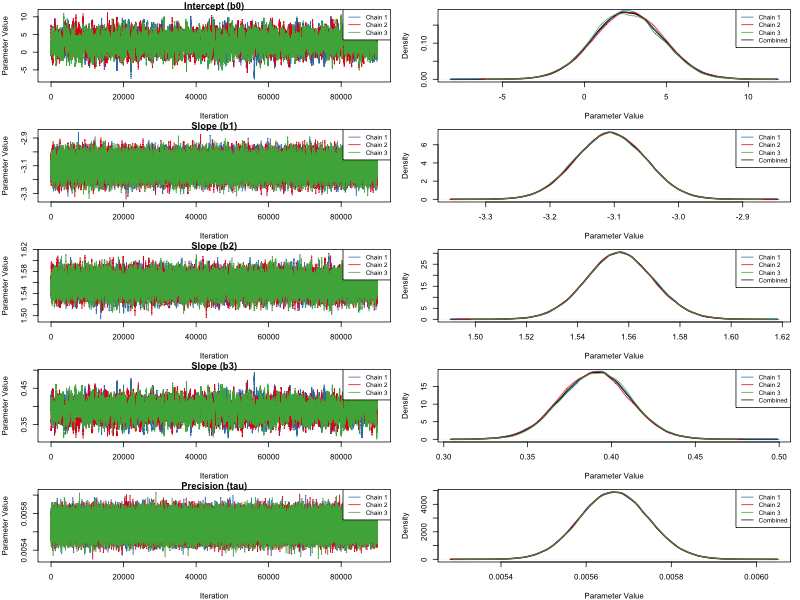 Figure 1. MCMC sampler of related dataset
Figure 1. MCMC sampler of related dataset
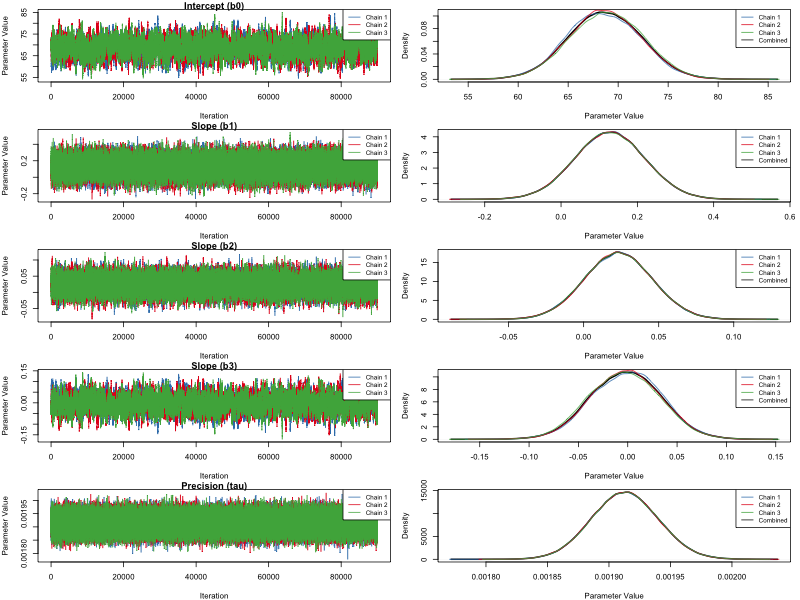 Figure 2. MCMC sampler of unrelated dataset
Figure 2. MCMC sampler of unrelated dataset